Abstract
Acute myeloid leukemia (AML) bearing mutations in the TP53 tumor suppressor gene is the most fatal of AMLs with a two-year overall survival between 0-15%. Patients with TP53 mutant (MT) AML typically respond to initial therapy but quickly relapse and die of disease due to chemotherapy resistance. We have undertaken to understand the mechanisms of chemotherapy resistance in TP53 MT AML. Initially, we purified AML blasts to 99% purity using flow sorting and performed RNA sequencing on TP53 MT and wildtype samples. Gene set enrichment analysis demonstrated that TP53 MT AML patient samples were enriched in the cholesterol synthesis or mevalonate pathway. This was validated using the BEAT AML dataset. Interestingly, p53 loss and/or mutation in breast, liver, colon and pancreatic cancer models leads to a dependency on an upregulated mevalonate pathway. Despite these data, the role of the mevalonate pathway in TP53 MT AML is unknown. We have subsequently explored the role of the mevalonate pathway in TP53 MT AML chemotherapy resistance. Experiments were performed in CRISPR-engineered TP53 mutated human AML cells with isogenic controls. Treatment with DNA damaging agents and analysis for cell survival confirmed that loss of wildtype p53 function enhanced cell survival. Recent results have demonstrated that chemotherapy resistance in AML cells is associated with an increase in oxidative phosphorylation (OXPHOS), which is dynamically induced by DNA damage. We confirmed that TP53 MT AML cells increase OXPHOS after DNA damage (Figure 1). Treatment of cells with rosuvastatin, a potent inhibitor of the mevalonate pathway, blocked this increase in OXPHOS and induced chemosensitivity. Lipidomic analysis demonstrates that the increase in OXPHOS correlates with an increase in coenzyme Q10 (CoQ10), a byproduct of the mevalonate pathway. Importantly, co-treatment of cells with a soluble form of mevalonate or a downstream byproduct, geranylgeranyl pyrophosphate (GGPP), recovered OXPHOS and subsequent chemoresistance. This indicates rosuvastatin's activity is specific to the mevalonate pathway, independent of cholesterol, and dependent on the GGPP branch of the pathway. Analysis is ongoing to determine the role of GGPP in regulating the mitochondrial response to DNA damage in TP53 MT cells, including if GGPP is required for CoQ10 synthesis as this remains a controversy in the literature. Overall, this data supports a model where TP53 MT AML cells dynamically upregulate the mevalonate pathway to regulate OXPHOS and avoid DNA damage-induced cell death (Figure 2). To determine if this model is physiologically relevant, we studied a patient derived xenograft (PDX) model of TP53 MT AML cells. Engrafted mice were treated with standard AML chemotherapeutic agent, cytarabine (AraC), for five days alone or in combination with high dose rosuvastatin. Interestingly, mice sacrificed three days after completing AraC administration continue to demonstrate an enhanced OXPHOS response. This response is blunted by co-treatment with rosuvastatin. As expected, TP53 MT AML cells respond modestly to AraC alone and this response was increased with the combination therapy. Overall, these results demonstrate that TP53 MT AML requires the mevalonate pathway for chemotherapy resistance and targeting of the mevalonate pathway in combination with chemotherapy may improve clinical responses. Through detailed biochemical analysis of this mechanism, we will elucidate novel therapeutic approaches to TP53 MT AML.
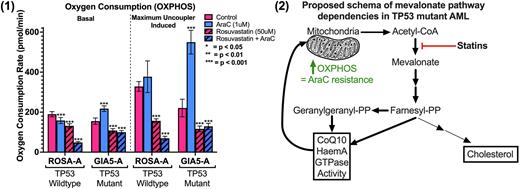
Figure Legend: (1) Oxygen consumption rate measured by seahorse technology in indicated cell lines and conditions. Statistics performed with unpaired T-test in GraphPad with stars indicating significance as per figure. (2) Proposed schema of mevalonate pathway dependencies in TP53 mutant (MT) AML. Bold arrows indicate increased byproduct dependency in TP53 MT AML cells treated with DNA damaging chemotherapy.
Disclosures
Carroll:Janssen Pharmaceuticals: Consultancy; Cartography Bioscences: Membership on an entity's Board of Directors or advisory committees.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal